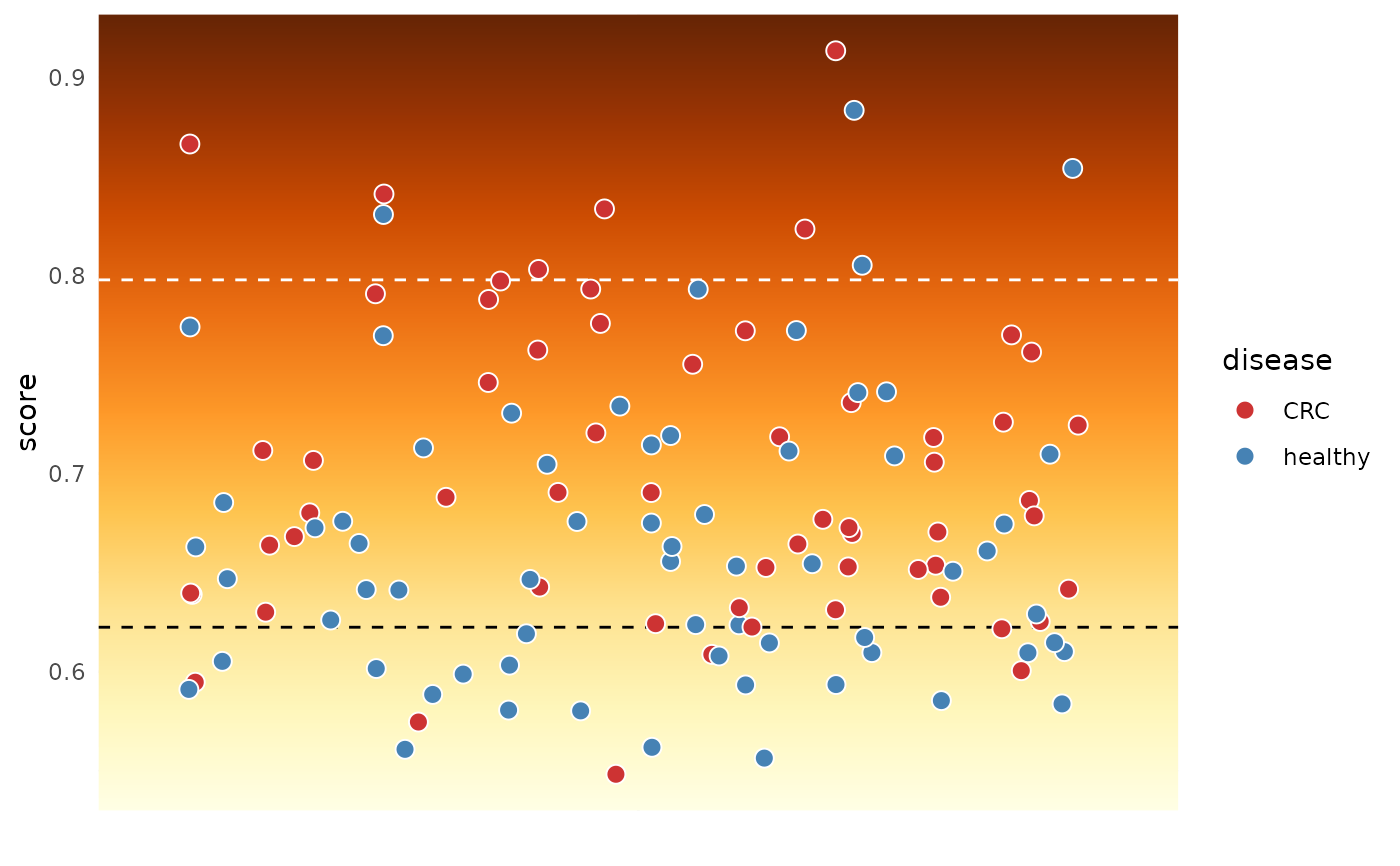
Dysbiosis Score Gradient Visualization
Arguments
- df
The data frame output from different dysbiosis calculators provided by dysbiosisR package.
- score
Variable to plot on x-axis.
- colors
colors to use for plotting. Default is NULL.
- show_points
Logical TRUE or FALSE.
- jitter_width
Passed to geom_jitter.
Examples
data("WirbelJ_2018")
library(RColorBrewer)
dist.mat <- phyloseq::distance(WirbelJ_2018, "bray")
# get reference samples
ref.samples <- sample_names(subset_samples(WirbelJ_2018,
disease == "healthy"))
dysbiosis_1 <- dysbiosisMedianCLV(WirbelJ_2018,
dist_mat = dist.mat,
reference_samples = ref.samples)
# get dysbiosis and normobiosis thresholds
dysbiosis_thres <- quantile(subset(dysbiosis_1, disease == "CRC")$score, 0.9)
normobiosis_thres <- quantile(subset(dysbiosis_1, disease == "CRC")$score, 0.1)
plotDysbiosisGradient(df=dysbiosis_1,
score="score",
high_line = dysbiosis_thres,
low_line = normobiosis_thres,
group_var = "disease",
group_colors=c("healthy" = "steelblue", "CRC"= "brown3"),
point_size = 2,
bg_colors = rev(brewer.pal(9, "YlOrBr")))